Artificial intelligence (AI) is changing the field of ophthalmology research, offering new methods for image analysis and use. The Artificial Intelligence-Ready Exploratory Atlas for Diabetes Insights (AI-READI) project aims to fill a critical gap in the lack of large, well-curated structured data sets for training AI models and to build a comprehensive, multimodal data set centered on type 2 diabetes mellitus (T2DM). Rather than focusing exclusively on T2DM pathology, the project incorporates a salutogenic lens, highlighting a variety of factors that contribute to health and resilience. This broader framework can help to understand T2DM, where physiologic, environmental, behavioral, psychological, and socioeconomic influences are intertwined. With retinal imaging as one of the major components of the AI-READI data set, the project is relevant to retinal researchers and clinicians exploring the intersection of diabetes, multidimensional health, and artificial intelligence.
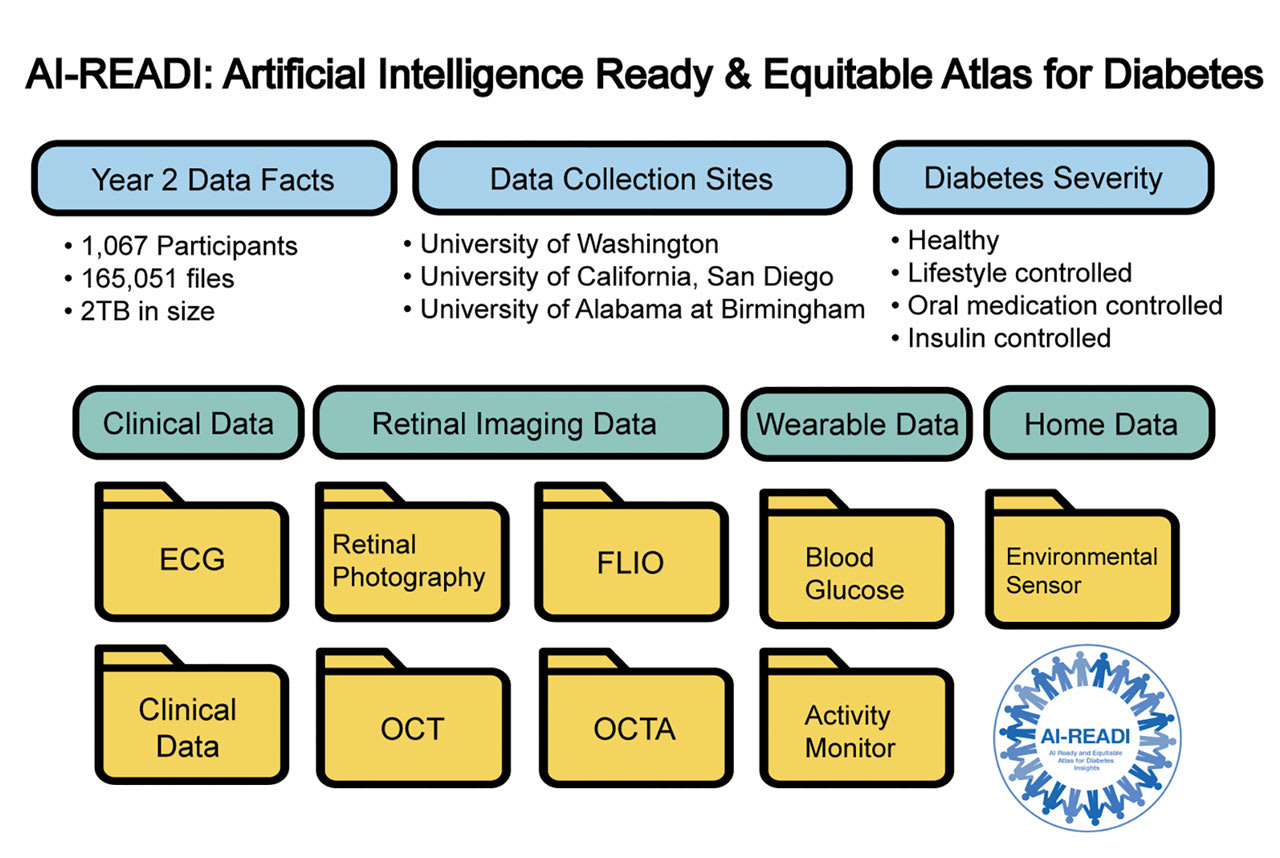
Figure 1. The AI-READI project aims to create and share a flagship dataset focused on type 2 diabetes. The data is optimized for future artificial intelligence and machine learning analyses that may provide critical insights into diabetes care. The figure shows the data elements included in the Year 2 release. Image courtesy Nayoon Gim, BS.
What is AI-READI
AI-READI is a data-generation project supported by the National Institutes of Health (NIH) through the Bridge2AI program. Its primary goal is to create a flagship, multimodal data set focused on type 2 diabetes.1,2 Over a 5-year period (2022-2026), the project aims to recruit 4,000 participants from 3 sites: the University of Washington, the University of Alabama at Birmingham, and the University of California, San Diego.3 Recruitment is strategically balanced across 4 categories of diabetes severity: individuals with no type 2 diabetes, those managing the condition through lifestyle changes, those on oral medications, and those using insulin.
A key feature of the AI-READI project is its commitment to data standardization, multimodality, and balance across cohorts. All data are processed following established standards within each modality, which is essential for interoperability and reproducibility—major challenges in developing AI systems. The data are shared annually via a web platform called fairhub.io, following FAIR principles (findable, accessible, interoperable, and reusable).4,5
Available Data
As of the year 2 release (v2.0.0 main study, released on November 8, 2024), data from 1,067 participants are available.4 The cohort includes 372 healthy individuals, 242 with prediabetes, 323 managing diabetes with oral medications, and 130 on insulin therapy. The entire data set includes 165,051 files and is around 2 TB in size. The data set spans more than 15 modalities and includes information from different phases of participant engagement: pre-visit, on-site visit, and post-visit monitoring (Figure 1).
Pre-visit data include self-reported surveys covering demographics, depression scale, Problem Areas in Diabetes questionnaire, diet, smoking history, alcohol use, marijuana use and vaping, social determinants of health, visual impairment, and access to eye care.
During on-site visits, a wide range of data are collected, including high-resolution retinal imaging using multiple techniques, such as color fundus photography, autofluorescence, infrared imaging, optical coherence tomography (OCT), OCT angiography (OCTA), and fluorescence lifetime imaging ophthalmoscopy (FLIO). Data were acquired using 7 imaging devices: Aurora (Optomed), Eidon (Centervue), Spectralis (Heidelberg Engineering), Cirrus (Carl Zeiss Meditec), Maestro2 (Topcon Healthcare), Triton (Topcon), and FLIO (Heidelberg). Additional on-site data include physical assessments (eg, height, weight, blood pressure, heart rate), blood tests (eg, HbA1c, lipid panel, C-peptide, troponin-T, NT-proBNP, CRP, CMP12), ECG, urine tests (albumin, creatinine), cognitive screening (MoCA), vision tests (lensometer, autorefraction, best-corrected visual acuity, letter contrast sensitivity), driving records (accident reports), monofilament testing for neuropathy, and biospecimen collection (blood samples for future RNA sequencing).1
Post-visit monitoring extends over 10 days and captures continuous glucose levels, wearable activity data, and environmental sensor readings. This includes real-time blood glucose data, physical activity metrics, and environmental exposure data such as temperature, humidity, particulate matter (PM1.0, PM4.0, PM10.0), nitrogen oxides (NOx), volatile organic compounds (VOCs), and light levels.
AI-READI collects biospecimens, such as blood and urine samples, for specific purposes. Plasma and serum are used for proteomics and metabolomics. Buffy coats are used to extract DNA for genomic studies, while PAXgene tubes preserve RNA for gene expression analysis. PBMCs are isolated from blood for immunologic research and iPSC generation. Urine is tested for kidney-related biomarkers. These data from biospecimens will be made available to researchers under future access policies.2
Standards Used for Each Data Type
The AI-READI project emphasizes the importance of using standardized data formats across all modalities to ensure data are interoperable, machine readable, and compatible with different analysis methods.
For imaging data, the DICOM (digital imaging and communications in medicine) standard is used. This includes 5 ophthalmic SOP (service-object pair) classes: ophthalmic photography, ophthalmic tomography, OCT B-scan volume analysis, height map segmentation, and OCT en face images.6 Each image is paired with rich metadata, such as participant ID, image dimensions, anatomic region, acquisition parameters, manufacturer details, and pixel-level data.
Electrocardiogram (ECG) data are collected using the Philips Pagewriter TC30 and stored using the WFDB (waveform database) standard. This consists of a .dat binary file containing waveform data and a .hea ASCII file that defines the data structure and includes annotations.
Environmental sensor data were collected using custom-designed sensor units placed in each participant’s home for 10 days. These sensors captured light levels, temperature, humidity, particulate matter, nitrogen oxides, and VOCs. Data are stored in ASCII format following the NASA Earth Science Data standard, a self-documenting tabular data format widely recognized for scientific use and readable by popular libraries like pandas in Python.7
Wearable activity monitor data were gathered using the Garmin Vivosmart 5 fitness tracker. During the 10-day monitoring period, this device recorded heart rate, oxygen saturation, sleep patterns, stress levels, respiration rate, and physical activity. The data files are provided in the mHealth JSON standard, which is both simple and human-readable, facilitating easy analysis.
Continuous glucose monitoring (CGM) data were collected using the Dexcom G6 device, which records glucose levels every 5 minutes. Participants wore the CGM device for 10 days. The resulting data are stored in JSON format, making it easy to access using standard data science libraries such as Python’s pandas or within Jupyter Notebooks. The data set also adheres to the mHealth JSON structure, a well-established framework for mobile health data.
Finally, clinical data are mapped using the OMOP (Observational Medical Outcomes Partnership) common data model, enabling standardized representation of medical records and enhancing compatibility with existing healthcare databases.8 Clinical lab tests, peripheral neuropathy, monofilament testing, physical assessment, questionnaires, cognition test (MoCA), and vision assessment data were mapped in the OMOP format and released as clinical data.
How to Access the Data
Access to the AI-READI data set is managed through a combination of public and controlled access mechanisms to ensure participant privacy while promoting open science. Public data are openly accessible for research purposes related to T2DM, which includes ECG, retinal imaging, clinical data, environmental sensor, wearable activity monitor, and continuous glucose monitor data. Data under controlled access includes sensitive information such as 5-digit ZIP codes, sex, race, ethnicity, genetic sequence data, past health records, medication history, and traffic and accident reports.
All data sets are hosted on FAIRhub, a cloud-based platform designed to help manage and share biomedical data sets in alignment with FAIR principles. The AI-READI data set can be accessed at https://fairhub.io/datasets/2. Comprehensive documentation for the data set is available at https://docs.aireadi.org.
The data set is free to use and available for download upon approval of a data access request, including for commercial use. Associated metadata are also available.9 To request access, researchers must authenticate via CILogon, confirm that their research is related to type 2 diabetes, and show they have been trained in research methods and ethics. Applicants must also submit a brief description of their research objectives and agree to the terms of the data set license. The AI-READI data set is released under a custom data license developed by the University of Washington, the data set’s licensor.10 An adequate data license is essential for enabling responsible data sharing because it helps ethical use and sets clear boundaries, while still allowing researchers to make meaningful use of the data. As data sharing becomes more central to scientific progress, thoughtful licensing will play an important role in balancing access with accountability. Users can then select the specific data types from the AI-READI data set they wish to access. Most requests are reviewed and approved within a few days, enabling timely access to this rich, multimodal data set.
Impact
By integrating diverse data modalities and applying modality specific standards. AI-READI establishes a new benchmark for developing AI ready data sets. The project not only advances opportunities for AI-driven innovation in diabetes research in general but also opens the door to novel applications in ophthalmic research. For retinal imaging specialists and researchers, the data set’s depth and breadth, particularly its multidevice ophthalmic imaging and relevant clinical data (eg, ophthalmic measurements and relevant questionnaires) offer a valuable resource for developing computational models aimed at understanding diabetic eye disease. RP
References
1. AI-READI Consortium. AI-READI: rethinking AI data collection, preparation and sharing in diabetes research and beyond. Nat Metab. 2024;6(12):2210-2212. doi:10.1038/s42255-024-01165-x
2. Owsley C, Matthies DS, McGwin G, et al. Cross-sectional design and protocol for Artificial Intelligence Ready and Equitable Atlas for Diabetes Insights (AI-READI). BMJ Open. 2025;15(2):e097449. doi:10.1136/bmjopen-2024-097449
3. Ferguson A, Drolet C, Kim T, et al. Artificial Intelligence Ready and Equitable Atlas for Diabetes Insights (AI-READI) study: report on recruitment strategy and pilot data collection. Invest Ophthalmol Vis Sci. 2024;65(7):3727-3727.
4. FAIRhub. Flagship data set of type 2 diabetes from the AI-READI project. November 8, 2024. Accessed April 1, 2025. https://fairhub.io/
5. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi:10.1038/sdata.2016.18
6. Gim N, Shaffer J, Owen J, et al. Streamlined DICOM standardization in retinal imaging: Bridging gaps in ophthalmic healthcare and AI research. Invest Ophthalmol Vis Sci. 2024;65(7):5879.
7. Shaffer J, Gim N, Wei R, Owen J, Lee CS, Lee AY. Portable environmental sensor enabling studies of exposome on ocular health. Invest Ophthalmol Vis Sci. 2024;65(7):6370.
8. Observational Health Data Sciences and Informatics (OHDSI). Standardized data: the OMOP common data model. Accessed April 7, 2025. https://www.ohdsi.org/data-standardization/
9. Patel B, Soundarajan S, Gasimova A, Gim N, Shaffer J, Lee AY. Clinical dataset structure: A universal standard for structuring clinical research data and metadata. Invest Ophthalmol Vis Sci. 2024;65(7):2418.
10. Contreras J, Evans B, Hurst S, et al. License terms for reusing the AI-READI dataset. Published online 2024. doi:10.5281/ZENODO.10642459










